Abstract
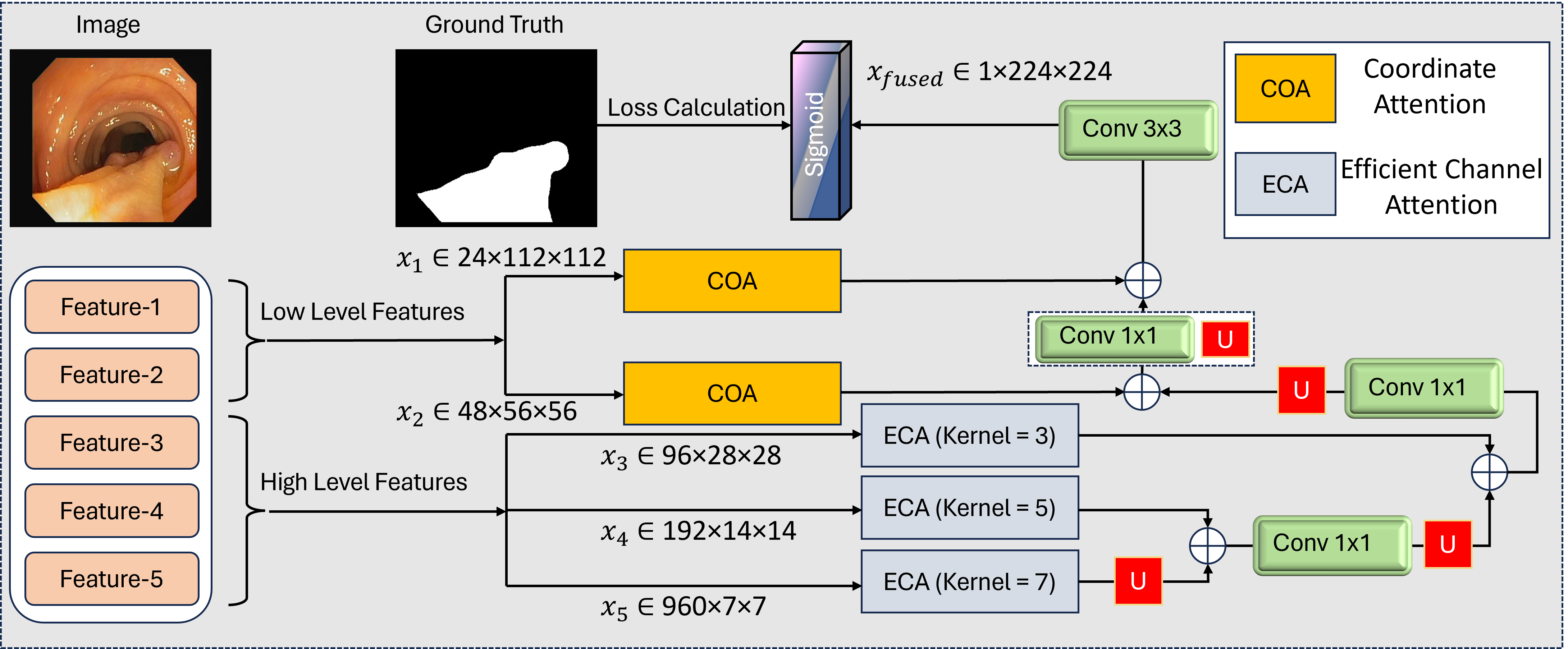
Accurate and real-time polyp segmentation plays a vital role in the early detection of colorectal cancer. However, existing methods often rely on computationally expensive backbones, single attention mechanisms, and suboptimal feature fusion strategies, limiting their practicality in real-world scenarios. In this work, we propose a lightweight yet effective deep learning framework that strikes a balance between precision and efficiency through a carefully designed architecture. Specifically, we adopt a MobileNetV4-based hybrid backbone to extract rich multi-scale features with significantly fewer parameters than conventional backbones, making the model well-suited for resource-constrained clinical settings. To enhance feature representation, we introduce a novel dual-attention guidance mechanism that integrates Efficient Channel Attention (ECA) for channel-wise refinement and Coordinate Attention (COA) for spatial modeling, which is particularly effective at delineating polyp boundaries. Additionally, we design a progressive multi-scale fusion strategy that hierarchically integrates feature maps from deep to shallow layers, preserving spatial details while enhancing contextual understanding. Extensive experiments on five benchmark polyp segmentation datasets demonstrate that our method consistently outperforms state-of-the-art approaches across both quantitative metrics and qualitative visualizations. Comprehensive ablation studies further validate the effectiveness of each component, highlighting the practical viability of our approach for real-time polyp segmentation applications.
Data Availability Statement
Data will be made available on request.
Funding
This work was supported without any funding.
Conflicts of Interest
The authors declare no conflicts of interest.
Ethical Approval and Consent to Participate
Not applicable.
Cite This Article
APA Style
Mohammed, E., Khan, A., Ullah, W., Khan, W., & Ahmed, M. J. (2025). Efficient Polyp Segmentation via Attention-Guided Lightweight Network with Progressive Multi-Scale Fusion. IECE Transactions on Intelligent Systematics, 2(2), 95–108. https://doi.org/10.62762/TIS.2025.389995
Publisher's Note
IECE stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Institute of Emerging and Computer Engineers (IECE) or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue