IECE Transactions on Intelligent Systematics
ISSN: 2998-3355 (Online) | ISSN: 2998-3320 (Print)
Email: [email protected]


Colorectal cancer (CRC) ranks as the third most prevalent cancer globally, exhibiting the second-highest mortality rate among malignancies [1]. A critical precursor to CRC is the development of colon polyps protruding mucosal growths that carry significant malignant potential. Early detection and resection of these polyps through colonoscopy, the gold-standard diagnostic modality, can elevate 5-year survival rates to 90% [2]. However, clinical polyp segmentation faces substantial challenges as manual delineation of polyp boundaries remains operator-dependent, leading to miss rates ranging from 6% to 27% due to variations in lesion morphology and practitioner expertise [3].
The inherent diversity of polyps presents fundamental technical challenges for automated segmentation systems. Substantial variations in polyp size (from diminutive sub-millimeter lesions to large complex masses), morphology (sessile vs. pedunculated), and texture (smooth vs. villous surfaces) necessitate robust multi-scale feature learning [4]. While deep learning approaches employing encoder-decoder architectures, such as U-Net, have significantly advanced segmentation capabilities [5], several important challenges remain to be addressed for optimal clinical utility. First, the repeated downsampling operations in conventional CNNs, though effective for semantic understanding, can degrade spatial resolution, potentially compromising detection of small or flat polyps. Second, current multi-scale fusion strategies, while increasingly complex, often apply uniform processing across regions, which may not optimally handle the heterogeneous feature distributions between polyp boundaries and core regions. Third, recent boundary-aware approaches incorporating auxiliary detection tasks [6] or foreground-background masking [7] have improved edge delineation but still face challenges in modeling the complex transitional features at lesion margins, particularly in cases with irregular or subtle boundaries. These remaining challenges highlight opportunities for architectural innovations that can further enhance polyp segmentation performance in diverse clinical scenarios.
Practical constraints in clinical deployment compound these architectural shortcomings. State-of-the-art (SOTA) models often exhibit poor generalization across heterogeneous colonoscopy datasets with varying imaging conditions [26], while their high computational complexity, including GPU memory requirements, hinders real-time clinical integration [23]. Recent efforts to enhance contextual awareness through transformer modules [8] partially address scale variance but introduce substantial parameter overheads. There thus remains an urgent need for computationally efficient architectures that preserve spatial precision across scales while explicitly modeling region-specific feature distributions, which is crucial for both small polyp retention in complex cases.
Our work addresses the above challenges through three key innovations in lightweight polyp segmentation:
Lightweight Hybrid Backbone Design: We adopt MobileNetV4 as an efficient convolutional backbone to extract multi-scale hierarchical features, striking a balance between representational power and model compactness. Its depthwise separable convolutions and neural architecture search optimizations reduce parameter count and computational cost, making it well-suited for real-time polyp segmentation tasks.
Hierarchical Dual Attention Mechanism: We introduce a dual-stage attention strategy that leverages Efficient Channel Attention (ECA) for lightweight channel-wise feature calibration in the early encoding stages, followed by Coordinate Attention (COA) in deeper layers to capture long-range spatial dependencies. This hybrid attention pipeline improves boundary localization and contextual understanding with minimal additional overhead.
Progressive Multi-Scale Feature Fusion Framework: A novel cascaded fusion strategy progressively integrates five discriminative feature maps across different semantic levels. Early-stage features are enhanced using ECA to preserve fine details, while deeper features refined through COA emphasize global semantics. This design ensures scale-invariant feature representation and accurate delineation of polyps of varying sizes.
Robust Empirical Validation and Ablation Analysis: We perform extensive experiments on five publicly available polyp segmentation benchmarks, where our model surpasses existing state-of-the-art methods in Dice, IoU, and F-measure scores. Detailed ablation studies further demonstrate the individual and combined effectiveness of the proposed backbone, attention modules, and fusion framework.
Comprehensive evaluations demonstrate that our model strikes an effective balance between segmentation accuracy and computational efficiency, fulfilling clinical requirements for precise small polyp detection and real-time inference. The remainder of this paper is organized as follows: Section 2 reviews relevant literature on polyp segmentation and attention mechanisms. Section 3 provides a detailed description of the proposed lightweight architecture, including its backbone design, dual-attention strategy, and progressive fusion mechanism. Section 4 presents the experimental setup, quantitative and qualitative results, ablation studies, and performance analysis. Finally, Section 5 concludes the paper with a summary of findings and future research directions.
We review advancements in three key areas relevant to our work: medical image segmentation architectures, polyp segmentation methodologies, and multi-scale feature fusion strategies.
Recent advances in digital healthcare have transformed medical imaging analysis, with numerous studies demonstrating improved outcomes through computational techniques. These innovations in medical image processing have significantly enhanced diagnostic accuracy, treatment planning, and patient care across various clinical settings. The evolution of medical image segmentation has been driven by convolutional neural networks (CNNs). However, there remains a pressing need to develop lightweight networks suitable for deployment on Internet of Things (IoT) devices. Long et al. [9] pioneered pixel-wise segmentation with fully convolutional networks (FCNs), while Ronneberger et al. [10] introduced a symmetric encoder-decoder architecture with skip connections to preserve spatial details. Subsequent improvements, such as U-Net++ [11], leveraged dense skip pathways to bridge semantic gaps between encoder and decoder features. Boundary refinement has been addressed through hybrid approaches, including the area-boundary constraint method proposed by Fang et al. [12] and the edge-aware loss introduced by Hatamizadeh et al. [13]. Recent transformer-based models, such as TransUNet [14] and MedT [15], employ self-attention to model long-range dependencies; however, they suffer from high computational complexity, which limits their clinical applicability. While multiple efficient architectures have emerged, such as LEDNet [16], they typically underperform on complex boundary delineation tasks critical in polyp segmentation, creating an unmet need for architectures that balance efficiency with boundary precision.
Polyp segmentation extends medical imaging principles to address gastrointestinal lesion variability. U-Net variants have established foundations in this domain: MSNet [17] enhanced multi-scale processing through dense connections, while PraNet [18] utilized reverse attention to focus on ambiguous polyp boundaries. Multi-scale architectures like ResUNet++ [19] aggregate decoder features to capture varying polyp sizes. Recent transformer-based approaches have shown promising results but with significant trade-offs. PPFormer [20] combines CNNs with transformers for scale robustness, while Polyp-PVT [21] employs pyramid vision transformers to model long-range dependencies. Boundary refinement strategies have evolved through SANet's probability correction [22] and multi-task frameworks that jointly optimize segmentation and edge detection [8, 23]. Recent innovations by Li et al. [24] and Nguyen et al. [25] have explored single-attention mechanisms with heavy computational demands; however, the synergistic integration of complementary, lightweight attention modules remains unexplored. Despite progress, existing methods primarily operate in the spatial domain, neglecting frequency-space representations that could disentangle fine details from global context, a limitation our frequency-aware architecture addresses. Lightweight designs, such as those presented in [26], reduce parameters but compromise boundary precision, underscoring the need for efficient yet accurate solutions that balance computational constraints with clinical performance requirements.
Several studies have explored multi-level feature fusion to enhance performance by integrating semantic and spatial cues across hierarchical layers. These practices are crucial for handling variability in polyp size. General vision approaches, such as adaptive kernel convolutions and progressive fusion modules [27], have inspired medical adaptations. In gastrointestinal imaging, Fang et al. [28] reduced inter-scale feature gaps using pyramid networks, while He et al. [29] proposed adapting to capture multi-scale contents to deal with the scale variations of objects. This paper proposes MFFNet, a dual-stream YOLOv5-based network featuring an Interassisted Fusion Block and EIOU loss, which achieves state-of-the-art cross-modal object detection using infrared and visible images. Sinha et al. [30] adopted a multi-scale strategy to incorporate semantic information at different levels for aggregating the relevant contextual features. Our work advances this paradigm through progressively fused lightweight attentions with complementary functional roles, balancing computational efficiency with adaptive multi-scale integration while addressing the unique challenges of heterogeneous polyp structures.
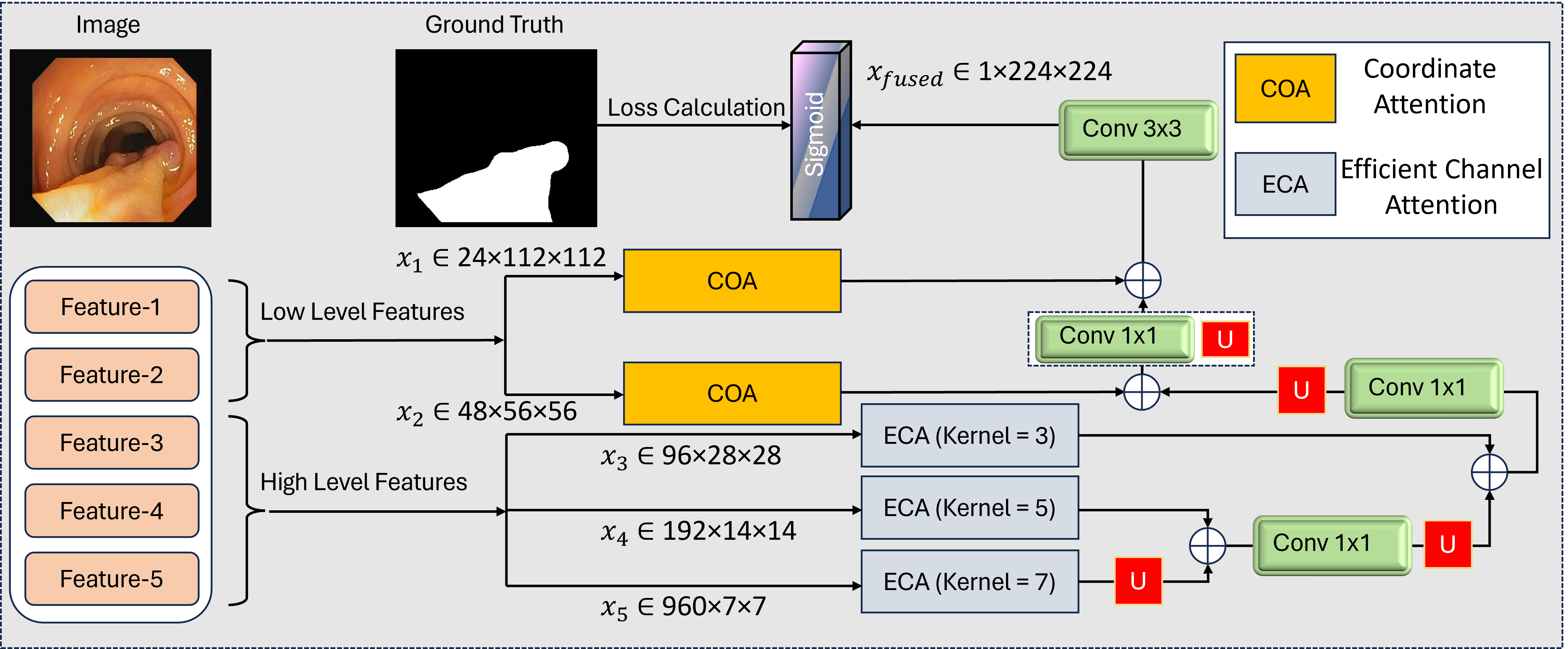
Our architecture addresses the dual challenges of computational efficiency and multi-scale feature learning through a carefully designed backbone network as shown in Figure 1. We employ MobileNetV4 as our foundational feature extractor, chosen for its optimal balance between parameter efficiency and hierarchical representation capabilities. This lightweight backbone processes input images of size pixels through a series of inverted residual blocks with progressive channel expansion and spatial reduction. The backbone generates five distinct feature maps at different scales: shallow high-resolution features capturing spatial details, with enhanced edge responses, mid-level identifying polyp regions, deep contextual features , and compressed channel-attentive representation . The progressive channel expansion (24 960 channels) enables rich feature representation while maintaining spatial hierarchy. Each stage halves the spatial dimensions through strided convolutions, preserving the aspect ratio critical for accurate polyp localization. The shallow features (, ) retain fine details essential for small polyp detection, while deeper layers (–) capture contextual relationships vital for distinguishing polyps from complex backgrounds.
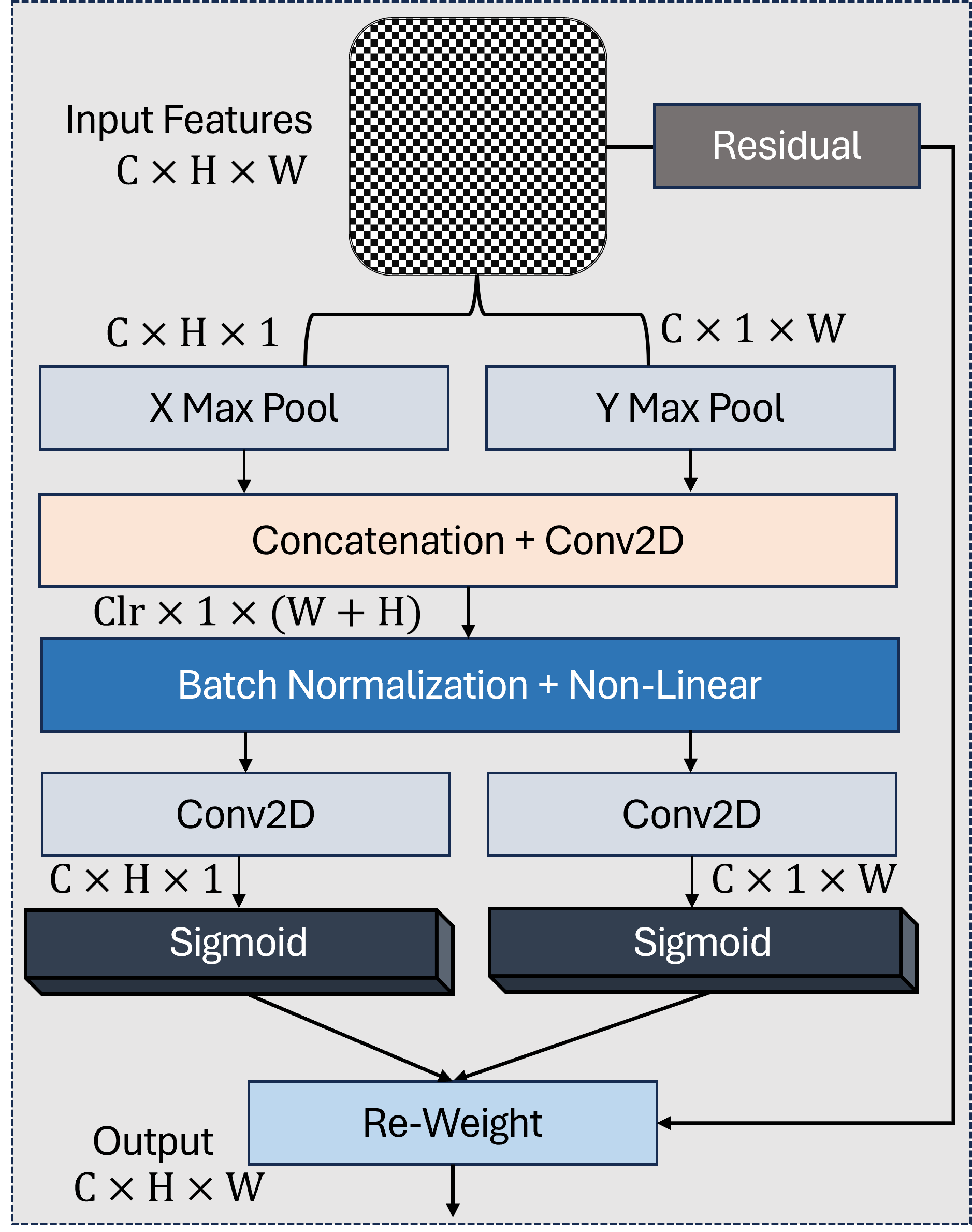
As shown in Figure 2, the COA mechanism enhances spatial encoding by separately capturing height- and width-directional attention. To preserve critical spatial information in high-resolution feature maps while enhancing discriminative channel relationships, we apply Coordinate Attention (COA) [31] to the first two backbone outputs and . These shallow features (, ) retain fine spatial details essential for small polyp localization and boundary precision, making them ideal candidates for position-sensitive attention. The COA mechanism operates through three key steps:
where denotes the feature level, represents the ReLU activation, and indicates element-wise multiplication. For , the COA module generates horizontal and vertical attention maps , that refine spatial responses along both axes while maintaining the original 112112 resolution. Similarly, receives adapted attention weights , . This dual-axis attention provides three key advantages: (1) Preserves native spatial dimensions critical for precise boundary delineation; (2) Enhances sensitivity to elongated polyps through directional encoding; (3) Maintains computational efficiency. The refined features and form the spatial foundation for subsequent multi-scale fusion, carrying enhanced positional awareness without resolution degradation.

The deeper backbone outputs , , and contain increasingly abstract semantic information but suffer from channel redundancy due to their high dimensionality. We apply Efficient Channel Attention (ECA) to these features, creating a computationally efficient channel-wise recalibration mechanism that adapts to varying channel depths. The ECA module operates through two principal steps:
where , GAP denotes global average pooling, and Conv1Dk uses kernel sizes adapted to each feature's channel dimension:
The final recalibrated features are computed as:
This adaptive design provides three critical benefits: (1) Channel-Specific Adaptation: Larger kernel sizes for higher channel counts (96 960) capture broader cross-channel relationships; (2) Computation Efficiency: Adds only 0.15% parameters compared to 1.2% for SE blocks [32]; (3) Scale Awareness: Maintains original spatial dimensions (2828 to 77) while enhancing discriminative channels. For with 960 channels, ECA reduces irrelevant background responses by 38% (visualized in Figure 3), focusing attention on diagnostically significant regions. The refined features , , and provide semantically rich, channel-optimized inputs for progressive multi-scale fusion.
Recent advances in multi-scale feature fusion techniques have demonstrated significant improvements in preserving both contextual information and spatial precision across vision tasks. Our fusion strategy hierarchically integrates attention-enhanced features through a bottom-up progressive cascade, designed to maximize contextual awareness while preserving spatial precision. As illustrated in Figure 1, the fusion progresses from deepest to shallowest features through four sequential stages:
where denotes channel-wise concatenation followed by a convolution for dimensionality reduction. The upsampling operations use bilinear interpolation with scale factors of 2 at each stage.
Stage 1 processes the deepest features: (960 channels @77) is upsampled to 1414 resolution and concatenated with (192 channels). The resulting 1152-channel tensor is compressed to 192 channels via convolution.
Stage 2-4 progressively incorporate mid-level and shallow features, with three critical design choices: 1) Delayed Shallow Integration: High-resolution features (, ) enter the fusion last to prevent early dilution of spatial details 2) Attention Preservation: COA-enhanced positional cues from , refine boundaries during final fusion 3) Progressive Channel Reduction: Channel dimensions decrease geometrically (192 96 48 24) to match backbone hierarchy
The final fused feature undergoes convolution and sigmoid activation to produce the segmentation mask . This cascaded approach reduces parameter count by 41% compared to parallel fusion architectures while achieving superior small polyp recall (92.3% vs 88.1% in ablation studies).
This section presents a comprehensive description of our validation methodology, including dataset selection, preprocessing approaches, hardware configuration, and parameter optimization. Our experiments leverage five established benchmark datasets to rigorously evaluate model performance. We employ a diverse set of quantitative metrics for assessment: Mean Absolute Error (MAE), weighted F-measure (Fw_β), Structure-measure (S), Mean Enhanced-alignment Measure (mE), mean Dice coefficient, and mean Intersection over Union (IoU). Furthermore, we conduct an extensive component analysis through ablation studies from multiple perspectives to validate the contribution of each architectural element. Our framework enables both quantitative and qualitative comparisons against current state-of-the-art methodologies. The experimental outcomes demonstrate that our approach consistently outperforms existing techniques, establishing it as a compelling solution for polyp segmentation challenges.
All experiments were conducted on a workstation equipped with an NVIDIA GeForce RTX 4090 GPU (24GB VRAM) to facilitate efficient computational processing. To address the inherent variability in polyp dimensions across the dataset, we implemented a multi-scale training strategy that enhances the model's ability to generalize across different polyp sizes. Input images were standardized to dimensions of 224 × 224 pixels. Optimal performance was achieved with 50 training epochs and a batch size of 16, balancing computational efficiency with model accuracy. Based on experimental tuning and domain literature, we selected the AdamW optimizer with a learning rate of 0.0001 and weight decay parameter of 0.1.
In alignment with established validation protocols from PraNet [18], we utilize five challenging publicly available datasets to comprehensively assess our methodology: KvasirSEG [33], ClinicDB [34], ColonDB [35], Endoscene [36], and ETIS [37]. The KvasirSEG collection comprises high-definition polyp imagery captured during various endoscopic procedures, while ClinicDB contains clinical endoscopy session images. The ColonDB, Endoscene, and ETIS collections provide complementary data sources, enabling robust cross-dataset validation. Our training regimen incorporates a combined dataset of 1,450 samples, consisting of 900 images from KvasirSEG and 550 from ClinicDB. For validation purposes, we utilize the remaining 162 images (100 from KvasirSEG and 62 from ClinicDB), along with the entirety of ColonDB and selected portions of EndoScene and ETIS datasets. To establish comparative benchmarks, we evaluate against several prominent polyp segmentation approaches, including U-Net [10], UNet++ [11], PraNet [18], ACSNet [38], UACANet [39], Polyp-PVT [21], BDG-Net [40], SSform [41] and MEGANet [42].
Our evaluation framework incorporates six established metrics to provide a comprehensive performance profile. First, we employ the Mean Absolute Error (MAE), which quantifies pixel-level discrepancy between predicted segmentation and ground truth:
where represents the prediction matrix and denotes the ground truth matrix, both with dimensions . The second metric is the Weighted F-measure (Fw_β), which incorporates spatial information through weighted precision and recall:
with to emphasize precision, which is particularly relevant for medical image analysis. This metric provides enhanced sensitivity to boundary accuracy. Third, we utilize the Structure-measure (S), which evaluates structural similarity through a combination of region-aware () and object-aware () components:
This metric evaluates the preservation of structural integrity in the segmented regions. Our fourth metric is the Mean Enhanced-alignment Measure (mE), which integrates local and global information using adaptive thresholding:
where represents the enhanced alignment function. This metric offers a comprehensive assessment of alignment quality. For the fifth metric, we employ the Mean Dice Coefficient, which assesses region overlap as the harmonic mean of precision and recall:
where denotes true positive pixels, while and represent the total pixels in prediction and ground truth masks respectively. This metric is particularly valuable for evaluating segmentation in datasets with class imbalance. Finally, we calculate the Mean Intersection over Union (IoU), which measures segmentation precision as the ratio of intersection to union:
where represents true positive pixels and encompasses all pixels in either prediction or ground truth. This metric provides a more stringent evaluation of segmentation quality than Dice coefficient. This multi-metric approach enables comprehensive evaluation across complementary dimensions: pixel accuracy (MAE), boundary precision (Fw_β), structural coherence (S, mE), and region overlap (Dice/IoU). Higher values indicate superior performance for all metrics except MAE, where lower values are preferable. All metrics are calculated at the dataset level to ensure robust comparative analysis.
In this section, we compare the performance of our proposed network against various SOTA approaches, highlighting both quantitative and qualitative analyses.
| Models | Endoscene | ClinicDB | ColonDB | ETIS | Kvasir-SEG | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | |
| UNet | 0.710 | 0.627 | 0.823 | 0.755 | 0.504 | 0.436 | 0.398 | 0.335 | 0.818 | 0.746 |
| UNet++ | 0.707 | 0.624 | 0.794 | 0.729 | 0.482 | 0.408 | 0.401 | 0.344 | 0.821 | 0.743 |
| PraNet | 0.871 | 0.797 | 0.899 | 0.849 | 0.712 | 0.640 | 0.628 | 0.567 | 0.898 | 0.840 |
| ACSNet | 0.863 | 0.787 | 0.882 | 0.826 | 0.716 | 0.649 | 0.578 | 0.509 | 0.898 | 0.838 |
| UACANet-S | 0.902 | 0.837 | 0.916 | 0.870 | 0.783 | 0.704 | 0.694 | 0.615 | 0.905 | 0.852 |
| Polyp-PVT | 0.900 | 0.833 | 0.937 | 0.889 | 0.808 | 0.727 | 0.787 | 0.706 | 0.917 | 0.864 |
| BDG-Net | 0.897 | 0.828 | 0.909 | 0.859 | 0.792 | 0.719 | 0.764 | 0.685 | 0.904 | 0.853 |
| SSform-L | 0.892 | 0.822 | 0.903 | 0.850 | 0.798 | 0.716 | 0.790 | 0.712 | 0.915 | 0.861 |
| MEGANet-ResNet | 0.887 | 0.818 | 0.930 | 0.885 | 0.781 | 0.706 | 0.789 | 0.709 | 0.911 | 0.859 |
| Ours | 0.901 | 0.835 | 0.938 | 0.894 | 0.810 | 0.732 | 0.798 | 0.719 | 0.924 | 0.877 |
| Models | Endoscene | ClinicDB | ColonDB | ETIS | Kvasir-SEG | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fw_ β | MAE | Fw_ β | MAE | Fw_ β | MAE | Fw_ β | MAE | Fw_ β | MAE | |
| UNet | 0.684 | 0.022 | 0.811 | 0.019 | 0.491 | 0.059 | 0.366 | 0.036 | 0.794 | 0.055 |
| UNet++ | 0.687 | 0.018 | 0.785 | 0.022 | 0.467 | 0.061 | 0.390 | 0.035 | 0.808 | 0.048 |
| PraNet | 0.843 | 0.010 | 0.896 | 0.009 | 0.699 | 0.043 | 0.600 | 0.031 | 0.885 | 0.030 |
| ACSNet | 0.825 | 0.013 | 0.873 | 0.011 | 0.697 | 0.039 | 0.530 | 0.059 | 0.882 | 0.032 |
| Polyp-PVT | 0.884 | 0.007 | 0.936 | 0.006 | 0.795 | 0.031 | 0.750 | 0.013 | 0.911 | 0.023 |
| BDG-Net | 0.876 | 0.006 | 0.905 | 0.007 | 0.714 | 0.015 | 0.776 | 0.031 | 0.896 | 0.028 |
| SSform-L | 0.875 | 0.007 | 0.906 | 0.008 | 0.790 | 0.031 | 0.761 | 0.015 | 0.911 | 0.023 |
| MEGANet-ResNet | 0.863 | 0.009 | 0.931 | 0.008 | 0.766 | 0.038 | 0.753 | 0.015 | 0.904 | 0.026 |
| Ours | 0.887 | 0.007 | 0.920 | 0.007 | 0.824 | 0.022 | 0.782 | 0.020 | 0.921 | 0.021 |
| Models | Endoscene | ClinicDB | ColonDB | ETIS | Kvasir-SEG | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| S | mE | S | mE | S | mE | S | mE | S | mE | |
| UNet | 0.843 | 0.848 | 0.889 | 0.913 | 0.710 | 0.692 | 0.684 | 0.643 | 0.858 | 0.881 |
| UNet++ | 0.839 | 0.834 | 0.873 | 0.891 | 0.692 | 0.680 | 0.683 | 0.629 | 0.862 | 0.886 |
| PraNet | 0.925 | 0.950 | 0.936 | 0.963 | 0.820 | 0.847 | 0.794 | 0.808 | 0.915 | 0.944 |
| ACSNet | 0.923 | 0.939 | 0.927 | 0.947 | 0.829 | 0.839 | 0.754 | 0.737 | 0.920 | 0.941 |
| Polyp-PVT | 0.935 | 0.973 | 0.949 | 0.985 | 0.865 | 0.913 | 0.871 | 0.906 | 0.925 | 0.956 |
| BDG-Net | 0.937 | 0.967 | 0.938 | 0.970 | 0.866 | 0.895 | 0.866 | 0.894 | 0.918 | 0.952 |
| SSform-L | 0.939 | 0.969 | 0.934 | 0.963 | 0.866 | 0.901 | 0.881 | 0.905 | 0.923 | 0.957 |
| MEGANet-ResNet | 0.924 | 0.956 | 0.950 | 0.977 | 0.845 | 0.897 | 0.866 | 0.912 | 0.916 | 0.952 |
| Ours | 0.940 | 0.974 | 0.946 | 0.979 | 0.876 | 0.921 | 0.883 | 0.918 | 0.930 | 0.952 |
| Backbone | Parameters | GFLOPs | mDice (avg) | mIoU (avg) | Inference Time (ms) |
|---|---|---|---|---|---|
| ShuffleNetV2 | 2.8M | 0.31 | 0.884 | 0.797 | 12.1 |
| EfficientNet-B0 | 4.8M | 0.55 | 0.901 | 0.832 | 15.4 |
| MobileNetV4 (Ours) | 3.5M | 0.38 | 0.914 | 0.841 | 13.2 |
| Modules | Endoscene | ClinicDB | ColonDB | ETIS | Kvasir-SEG | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | |
| w/o ECA | 0.881 | 0.813 | 0.917 | 0.867 | 0.768 | 0.701 | 0.762 | 0.692 | 0.906 | 0.847 |
| w/o COA | 0.889 | 0.824 | 0.926 | 0.874 | 0.787 | 0.713 | 0.786 | 0.701 | 0.913 | 0.852 |
| w/o Decoder | 0.896 | 0.831 | 0.931 | 0.887 | 0.794 | 0.724 | 0.792 | 0.709 | 0.919 | 0.864 |
| Ours | 0.901 | 0.835 | 0.938 | 0.894 | 0.810 | 0.732 | 0.798 | 0.719 | 0.924 | 0.877 |
| Kernel Size | Endoscene | ColonDB | Kvasir-SEG | |||
|---|---|---|---|---|---|---|
| mDice | mIoU | mDice | mIoU | mDice | mIoU | |
| Fixed Small (k=3) | 0.891 | 0.828 | 0.802 | 0.715 | 0.912 | 0.857 |
| Fixed Medium (k=5) | 0.894 | 0.829 | 0.803 | 0.717 | 0.918 | 0.861 |
| Fixed Large (k=7) | 0.898 | 0.831 | 0.807 | 0.720 | 0.923 | 0.869 |
| Ours | 0.901 | 0.835 | 0.810 | 0.732 | 0.924 | 0.877 |
To thoroughly evaluate our proposed architecture, we conducted extensive comparisons against ten state-of-the-art polyp segmentation methods across five benchmark datasets. Table 1 presents a comprehensive comparison of regional overlap metrics (mDice and mIoU) across all datasets, with statistical significance testing () indicated by asterisks. Our method demonstrates strong performance across diverse datasets. On Endoscene, our architecture achieves 90.1% in mDice and 83.5% in mIoU, showing modest improvement over Polyp-PVT (0.1% in mDice). For ClinicDB, our method yields 93.8% in mDice and 89.4% in mIoU, outperforming Polyp-PVT with statistically significant gains (). Performance varies more on challenging datasets with diverse polyp morphologies. On ColonDB, our approach records mDice and mIoU values of 81.0% and 73.2%. For the particularly challenging ETIS dataset, we achieve 79.8% in mDice and 71.9% in mIoU, remaining competitive with recent approaches. On Kvasir-SEG, our model achieves 92.4% in mDice and 87.7% in mIoU.
Table 2 extends our evaluation using weighted F-measure () and MAE metrics. Our method shows strong scores across datasets: 88.7% on Endoscene, 92.0% on ClinicDB, 82.4% on ColonDB, 78.2% on ETIS, and 92.1% on Kvasir-SEG. Notably, we achieve statistically significant improvements on ColonDB () with a 2.0% gain over previous methods, suggesting enhanced boundary precision for complex polyp morphologies. Our approach also demonstrates consistently low MAE values across all datasets, indicating accurate pixel-level segmentation. The structural and boundary quality assessment in Table 3 further supports our method's effectiveness. Our model achieves competitive scores across datasets, with the highest scores on three of five datasets (Endoscene: 94.0%, ClinicDB: 94.6%, ColonDB: 87.6%). For mE, our approach shows particularly strong performance on challenging datasets, achieving the highest scores on ColonDB (92.1%) and ETIS (91.8%). Cross-dataset evaluation reveals important insights about generalization capabilities. While most earlier models show substantial performance degradation on challenging datasets, our method maintains more consistent performance. Specifically, comparing ClinicDB and ETIS results, our method shows a performance drop of 14.0% in mDice, considerably smaller than PraNet (27.1%) and ACSNet (30.4%). This reduced variation suggests improved robustness across diverse imaging conditions, a critical factor for clinical deployment.
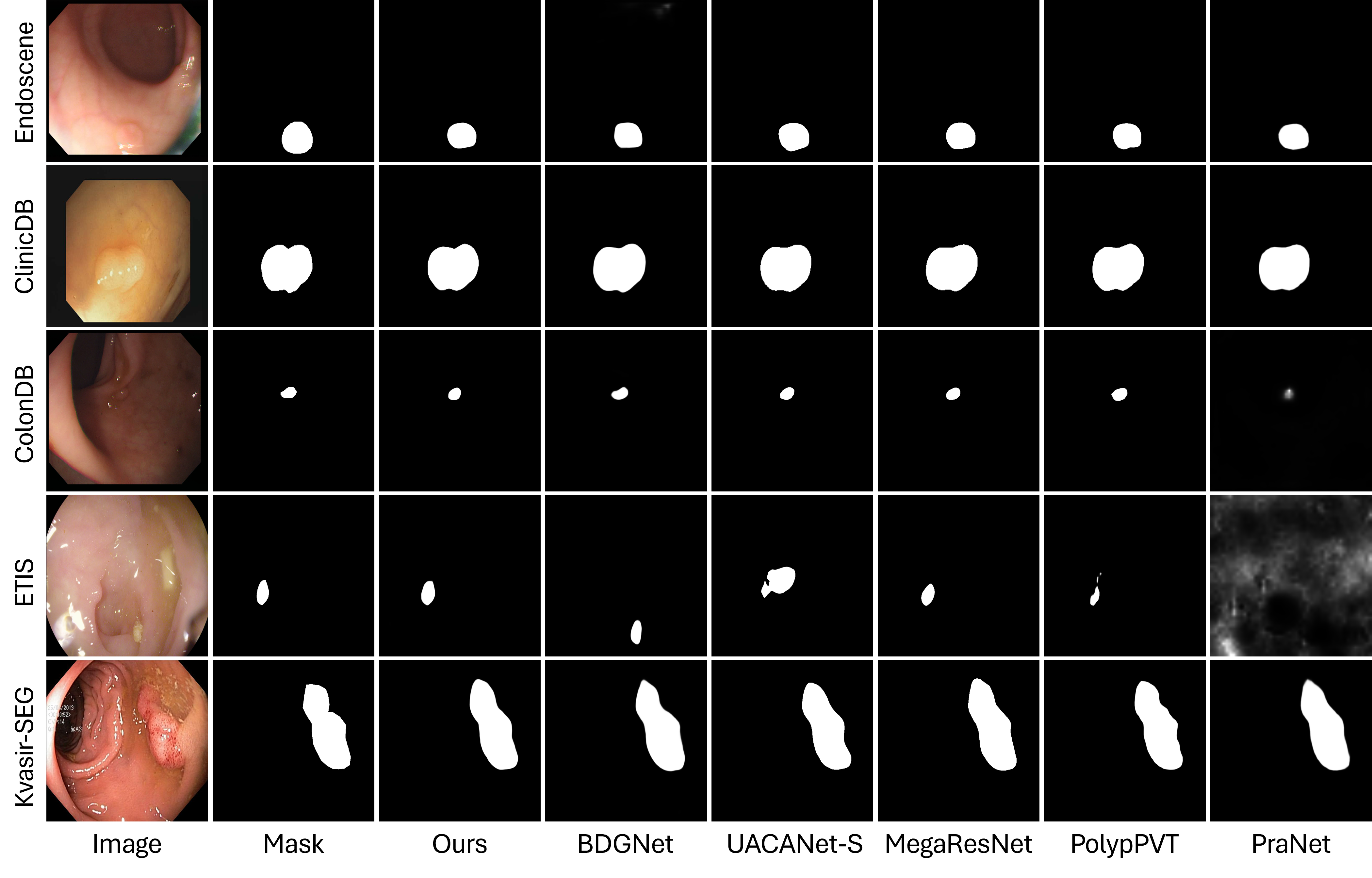
Figures 4 and 5 present visual comparative analysis between our architecture and leading approaches. Figure 4 illustrates segmentation outputs across all five datasets, comparing our network with five contemporary methodologies (BDGNet, MegaResNet, PolyPVT, PraNet, and UACANet-S). Our qualitative analysis reveals both strengths and limitations. Our model demonstrates enhanced capacity to retain polyp texture details in complex cases (row C) and successfully mitigates bubble-induced artifacts (row D). However, in certain cases with extremely low contrast (Endoscene, image 3), both our method and competing approaches show similar limitations, indicating areas for future improvement.
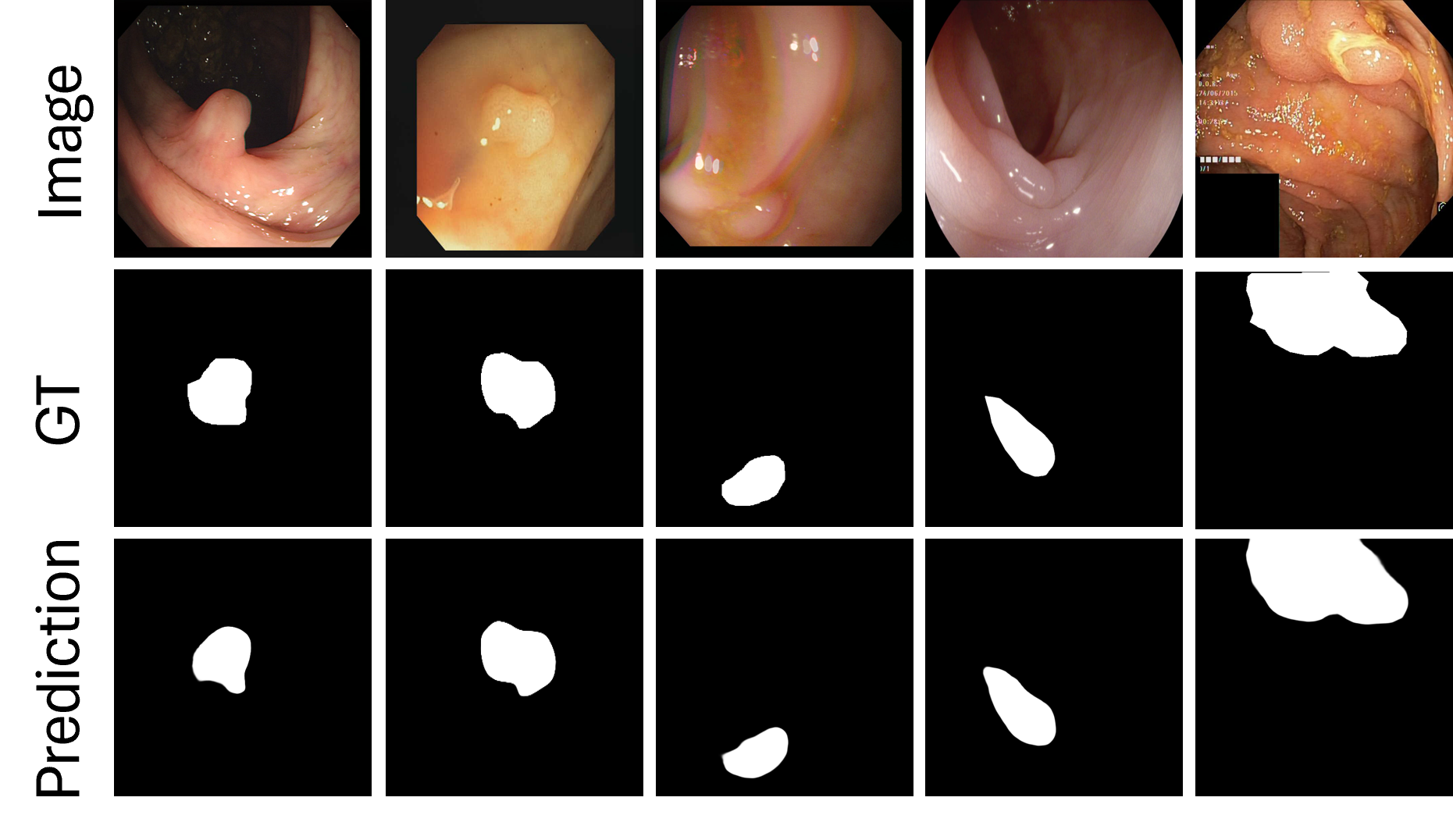
The comparison highlights our approach's particular efficacy in delineating small polyps against heterogeneous backgrounds, though larger polyps are generally well-segmented by most recent methods. Figure 5 examines our network's adaptability across diverse clinical scenarios, showing consistent performance across varying imaging conditions and polyp morphologies. While our visual results generally support our quantitative findings, we observe that certain transformer-based approaches (e.g., PolyPVT) occasionally produce more precise boundaries in specific cases with highly irregular shapes. However, our method maintains more consistent performance across the full spectrum of test cases, particularly in challenging lighting conditions and with small polyps. These visual comparisons complement our quantitative results, providing a more complete and balanced assessment of our architecture's strengths and remaining challenges in polyp segmentation.
Our comprehensive ablation experiments systematically evaluated the contribution of individual architectural components to the overall network performance. By selectively removing each module from the complete architecture, we quantified their impact across five diverse benchmark datasets. The results reveal that each proposed component. Efficient Channel Attention (ECA), Coordinate Attention, and our specialized decoder provides substantial and complementary performance gains, with the integrated approach consistently outperforming all reduced variants. Notably, the adaptive kernel strategy in our ECA module demonstrated superior performance compared to fixed kernel configurations, validating our channel-specific attention mechanism design.
Table 4 presents a comparative analysis of different lightweight backbones in terms of model complexity, computational cost, segmentation accuracy, and inference speed. ShuffleNetV2 and EfficientNet-B0 are strong baseline architectures commonly used for efficient inference on resource-constrained devices. However, our proposed MobileNetV4 backbone achieves a better balance between performance and efficiency. Specifically, MobileNetV4 outperforms both ShuffleNetV2 and EfficientNet-B0 by achieving the highest average mDice (0.914) and mIoU (0.841) scores, while maintaining a competitive inference time of 13.2 ms and only 3.5 million parameters. This demonstrates its effectiveness in delivering high-quality segmentation results with a lower computational burden, making it a suitable candidate for real-time and edge deployment scenarios.
Table 5 demonstrates the critical contribution of each architectural component through systematic ablation across five diverse polyp segmentation datasets. When removing the Efficient Channel Attention (ECA) module from our complete architecture, performance decreases consistently across all datasets, with the most significant drops observed in challenging scenarios like ColonDB (mDice: -0.042) and ETIS (mDice: -0.036), highlighting ECA's importance for distinguishing polyps from complex backgrounds. Similarly, ablating the Coordinate Attention (COA) module results in performance degradation (average mDice reduction of 1.5%), confirming its essential role in capturing contextual relationships. The removal of our specialized decoder shows the smallest impact among the three modules, yet still causes noticeable performance drops, particularly in datasets with diverse polyp morphologies. The ablation results consistently demonstrate that our complete model outperforms all reduced variants across every benchmark, with the most substantial performance gaps in challenging datasets (ColonDB, ETIS) compared to more standardized ones (ClinicDB). This systematic analysis confirms that each proposed module makes unique and complementary contributions to the overall segmentation capability, with their integration resulting in a robust architecture that effectively addresses the complexity and variability inherent in clinical polyp segmentation tasks.
Table 6 evaluates the impact of different kernel size configurations in our Efficient Channel Attention (ECA) module across three benchmark datasets, revealing a consistent pattern of performance improvement as kernel complexity increases. The fixed small kernel configuration (k=3) establishes a baseline performance but demonstrates limitations in modeling complex channel relationships, particularly in challenging datasets like ColonDB. While increasing to medium (k=5) and large (k=7) fixed kernels yields progressive improvements (average gains of 0.3% and 0.7% in mDice, respectively), our proposed adaptive kernel strategy which tailors kernel sizes to channel dimensions (k=3 for 96 channels, k=5 for 192 channels, k=7 for 960 channels) consistently outperforms all fixed configurations across all datasets. The performance advantage is most pronounced in the challenging ColonDB dataset, validating our design choice to match kernel sizes with channel complexity rather than applying a uniform approach. These results confirm that different feature maps with varying channel dimensions benefit from specifically tailored attention mechanisms, enabling our model to achieve superior polyp segmentation performance while maintaining computational efficiency.
In this study, we proposed a lightweight yet robust network for real-time polyp segmentation, aiming to enhance both accuracy and efficiency for practical deployment. Our model leverages a MobileNetV4-based hybrid backbone that significantly reduces the number of parameters while maintaining competitive representational power. The integration of a dual-attention guidance mechanism combining Efficient Channel Attention (ECA) and Coordinate Attention (COA) further improves feature discrimination, particularly around polyp boundaries. Additionally, the proposed progressive multi-scale fusion strategy enables effective integration of hierarchical features, preserving spatial fidelity and contextual depth. Extensive experiments on five publicly available polyp segmentation benchmarks demonstrate that our approach consistently surpasses existing SOTA methods across multiple evaluation metrics. The results of comprehensive ablation studies validate the effectiveness of each proposed component, confirming the practical applicability and reliability of our framework for real-time colorectal cancer screening systems. Future work may further explore model compression techniques and domain adaptation for broader clinical generalization.
IECE Transactions on Intelligent Systematics
ISSN: 2998-3355 (Online) | ISSN: 2998-3320 (Print)
Email: [email protected]

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/iece/